Industries
Compute on Dynex
DYNEX CORE
Tech Primer
Industries
Compute on Dynex
DYNEX CORE
Tech Primer
Industries
Compute on Dynex
DYNEX CORE
Tech Primer
Pharmaceutical
Pharmaceutical
Pharmaceutical

Quantum computing is on the verge of transforming the pharmaceutical industry by vastly improving the speed and accuracy at which companies can discover and develop new drugs. This cutting-edge technology offers the ability to simulate and analyze complex molecular interactions at a quantum level, something that traditional computing methods struggle to accomplish due to the sheer complexity of biological systems. By enabling more precise modeling of molecular structures and their interactions with biological targets, quantum computing can significantly reduce the time and cost associated with the drug discovery process, allowing for the rapid identification of promising compounds.
Furthermore, quantum computing has the potential to revolutionize personalized medicine. It can process vast datasets from genetic information, enabling the design of treatments that are tailored to individual genetic profiles, thereby enhancing their effectiveness and reducing side effects. The ability to quickly analyze and interpret large volumes of genetic data also accelerates the understanding of diseases at a molecular level, leading to breakthroughs in the treatment of complex conditions such as cancer, Alzheimer's, and heart disease.
In addition to drug discovery and personalized medicine, quantum computing can optimize manufacturing processes and supply chain logistics in the pharmaceutical industry. By identifying the most efficient pathways for chemical synthesis, quantum computing can help in the production of drugs at lower costs and with reduced environmental impact. Similarly, sophisticated quantum algorithms can improve supply chain efficiency, ensuring that medications are produced and distributed in the most effective manner possible.
As quantum computing technology matures and becomes more accessible, its integration into the pharmaceutical industry is poised to usher in a new era of medical innovation. The promise of faster, more efficient drug discovery and development, coupled with the potential for personalized treatments, positions quantum computing as a key driver of future advancements in healthcare and medicine, marking a significant leap forward in our ability to combat disease and improve human health.
Drug Repurposing With 3D Molecular Quantum Methods
From concept to treating a patient, it can take 10 years for a single treatment. Drug repositioning, repurposing, re-tasking, re-profiling or drug rescue is the process by which approved drugs are employed to treat a disease they were not initially intended/designed for. Virtual screening has become essential at the early stages of drug discovery. However, the process still typically takes a long time to execute since it generally relies on measuring chemical similarities among molecules. Even for today’s processors, this exercise comprises a major challenge since it is computationally heavy and expensive. Accordingly, most of the well-known methods typically use 2D molecular fingerprints to include structural information that represents substructural characteristics of molecules as vectors. These methods do not take into consideration relevant aspects of molecular structures such as 3D folding, although they are efficient in terms of execution times. At the expense of higher computing times, considering 3D structural properties of molecules substantially increases the accuracy of results. The 3D molecular Quantum method is computed efficiently on the Dynex platform and provides a superior virtual screening method.

Scientific background: Drug repurposing based on a quantum-inspired method versus classical fingerprinting uncovers potential antivirals against SARS-CoV-2, Jimenez-Guardeño JM, Ortega-Prieto AM, Menendez Moreno B, Maguire TJA, Richardson A, Diaz-Hernandez JI, et al. (2022); PLoS Comput Biol 18(7): e1010330

RNA Folding
Finds the optimal stem configuration of the RNA sequence from the HIV virus and the Tobacco Mild Green Mosaic Virus using the Dynex platform. The example takes an RNA sequence and applies a quadratic model in pursuit of the optimal stem configuration.
> Jupyter Notebook
Scientific background: Fox DM, MacDermaid CM, Schreij AMA, Zwierzyna M, Walker RC. RNA folding using quantum computers,. PLoS Comput Biol. 2022 Apr 11;18(4):e1010032. doi: 10.1371/journal.pcbi.1010032. PMID: 35404931; PMCID: PMC9022793
Enzyme-Target Prediction on the Dynex Platform
The Dynex SDK based program predicts potential interactions between enzymes and target molecules and leverages the principles of quantum mechanics.
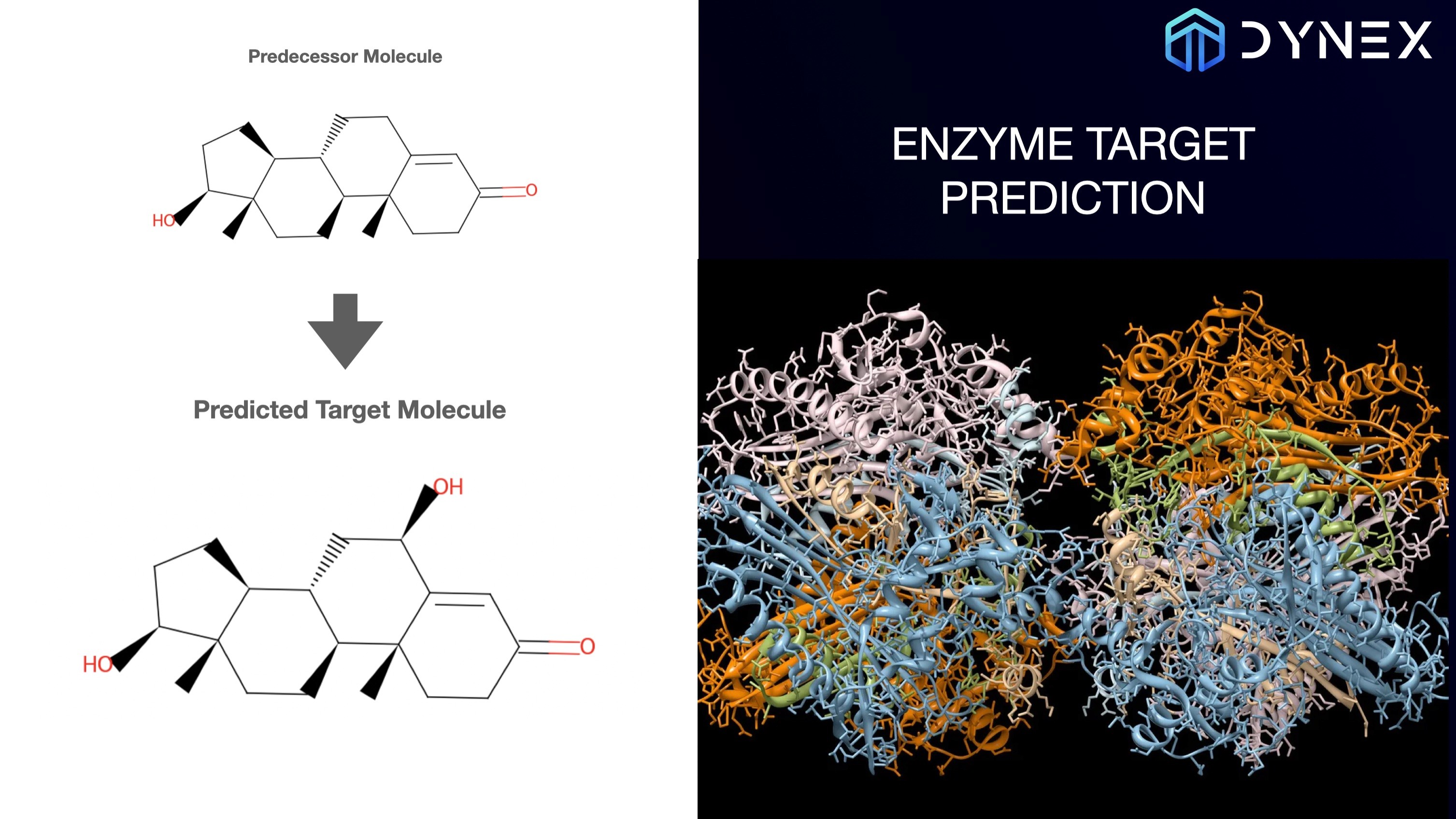
> Jupyter Notebook
Scientific background: Hoang M Ngo, My T Thai, Tamer Kahveci, QuTIE: Quantum optimization for Target Identification by Enzymes, Bioinformatics Advances, 2023;, vbad112
Efficient Exploration of Phenol Derivatives with Dynex
Molecule screening from a vast number of possible compounds is a challenging task. The emergence of quadratic unconstrained binary optimization (QUBO) solvers provides alternatives to address this issue. We propose a process for screening molecules by integrating QUBO solvers and density functional theory (DFT) calculations. As a proof-of-concept work, we map the problem of screening phenolic inhibitors onto the QUBO model. We approximate the bond dissociation energy (BDE) of the −OH bond, an indicator of good polymeric inhibitors, into the QUBO model by modifying the group contribution method (GCM) with the aid of DFT calculations. We demonstrate a strong correlation between this QUBO model and the data from DFT, with the correlation coefficient and Spearman’s coefficient of 0.82 and 0.86, respectively, when tested on the 85 given molecules. This mapping allows us to identify the candidates through the QUBO solver, whose BDEs are validated through DFT calculations, as well. Our work provides a promising direction for incorporating the GCM into QUBO solvers to tackle the molecule screening problems.
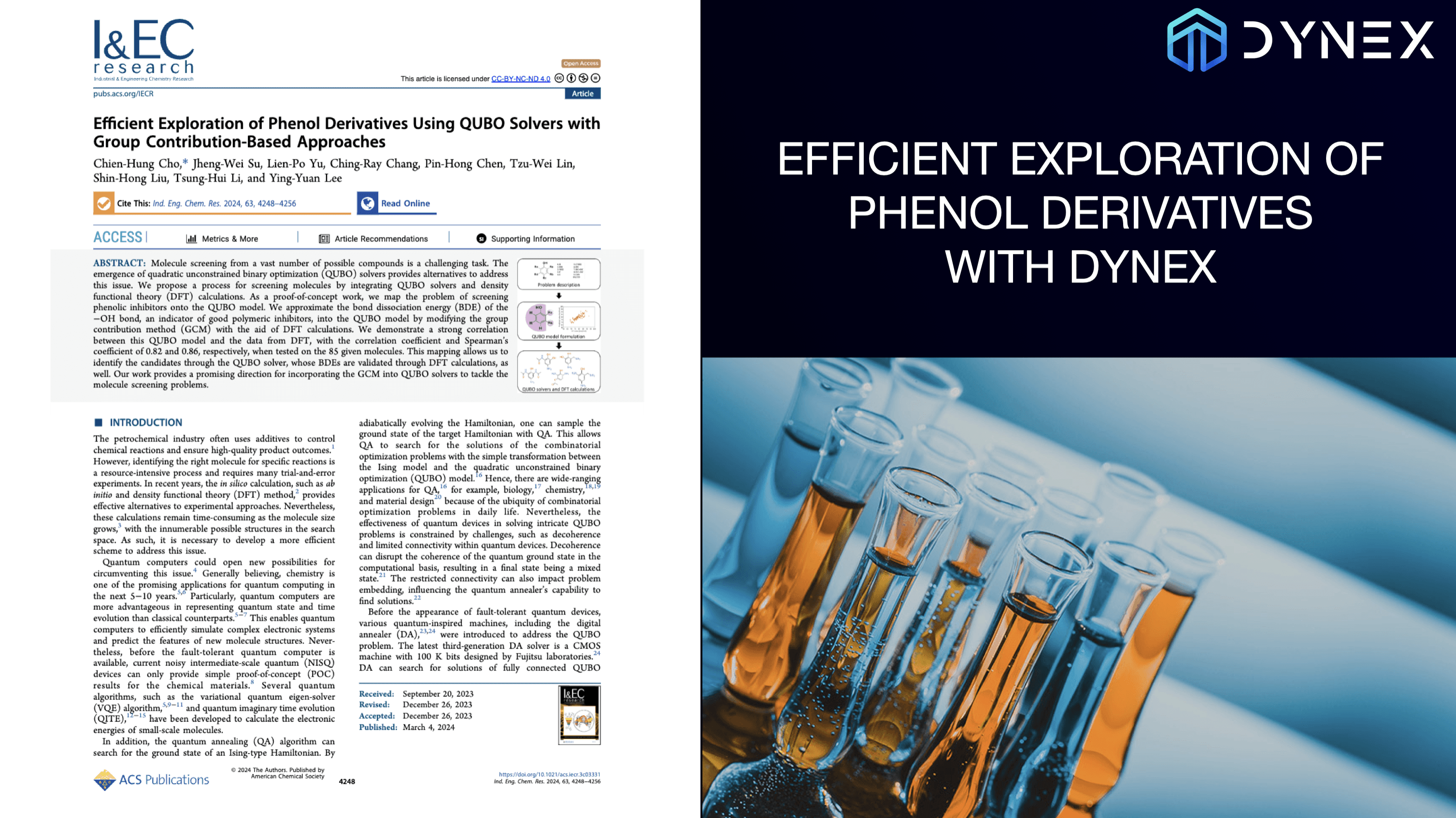
Scientific background: Efficient Exploration of Phenol Derivatives Using QUBO Solvers with Group Contribution-Based Approaches; Chien-Hung Cho, Jheng-Wei Su, Lien-Po Yu, Ching-Ray Chang, Pin-Hong Chen, Tzu-Wei Lin, Shin-Hong Liu, Tsung-Hui Li, and Ying-Yuan Lee; Industrial & Engineering Chemistry Research 2024 63 (10), 4248-4256; DOI: 10.1021/acs.iecr.3c03331
Breast Cancer Prediction
This examples shows using the Dynex SDK Scikit package which provides a scikit-learn transformer for feature selection using the Dynex Neuromorphic Computing Platform. The number of features have impact on neural network training and accuracy. It will be demonstrated how a significant reduction of features lead to similar (or even better) results.
> Jupyter Notebook
Scientific background: Bhatia, H.S., Phillipson, F. (2021). Performance Analysis of Support Vector Machine Implementations on the D-Wave Quantum Annealer. In: Paszynski, M., Kranzlmüller, D., Krzhizhanovskaya, V.V., Dongarra, J.J., Sloot, P.M.A. (eds) Computational Science – ICCS 2021. ICCS 2021. Lecture Notes in Computer Science(), vol 12747. Springer, Cham
> Automotive, Aerospace, Super-Sports and Space

Quantum computing is on the verge of transforming the pharmaceutical industry by vastly improving the speed and accuracy at which companies can discover and develop new drugs. This cutting-edge technology offers the ability to simulate and analyze complex molecular interactions at a quantum level, something that traditional computing methods struggle to accomplish due to the sheer complexity of biological systems. By enabling more precise modeling of molecular structures and their interactions with biological targets, quantum computing can significantly reduce the time and cost associated with the drug discovery process, allowing for the rapid identification of promising compounds.
Furthermore, quantum computing has the potential to revolutionize personalized medicine. It can process vast datasets from genetic information, enabling the design of treatments that are tailored to individual genetic profiles, thereby enhancing their effectiveness and reducing side effects. The ability to quickly analyze and interpret large volumes of genetic data also accelerates the understanding of diseases at a molecular level, leading to breakthroughs in the treatment of complex conditions such as cancer, Alzheimer's, and heart disease.
In addition to drug discovery and personalized medicine, quantum computing can optimize manufacturing processes and supply chain logistics in the pharmaceutical industry. By identifying the most efficient pathways for chemical synthesis, quantum computing can help in the production of drugs at lower costs and with reduced environmental impact. Similarly, sophisticated quantum algorithms can improve supply chain efficiency, ensuring that medications are produced and distributed in the most effective manner possible.
As quantum computing technology matures and becomes more accessible, its integration into the pharmaceutical industry is poised to usher in a new era of medical innovation. The promise of faster, more efficient drug discovery and development, coupled with the potential for personalized treatments, positions quantum computing as a key driver of future advancements in healthcare and medicine, marking a significant leap forward in our ability to combat disease and improve human health.
Drug Repurposing With 3D Molecular Quantum Methods
From concept to treating a patient, it can take 10 years for a single treatment. Drug repositioning, repurposing, re-tasking, re-profiling or drug rescue is the process by which approved drugs are employed to treat a disease they were not initially intended/designed for. Virtual screening has become essential at the early stages of drug discovery. However, the process still typically takes a long time to execute since it generally relies on measuring chemical similarities among molecules. Even for today’s processors, this exercise comprises a major challenge since it is computationally heavy and expensive. Accordingly, most of the well-known methods typically use 2D molecular fingerprints to include structural information that represents substructural characteristics of molecules as vectors. These methods do not take into consideration relevant aspects of molecular structures such as 3D folding, although they are efficient in terms of execution times. At the expense of higher computing times, considering 3D structural properties of molecules substantially increases the accuracy of results. The 3D molecular Quantum method is computed efficiently on the Dynex platform and provides a superior virtual screening method.

Scientific background: Drug repurposing based on a quantum-inspired method versus classical fingerprinting uncovers potential antivirals against SARS-CoV-2, Jimenez-Guardeño JM, Ortega-Prieto AM, Menendez Moreno B, Maguire TJA, Richardson A, Diaz-Hernandez JI, et al. (2022); PLoS Comput Biol 18(7): e1010330

RNA Folding
Finds the optimal stem configuration of the RNA sequence from the HIV virus and the Tobacco Mild Green Mosaic Virus using the Dynex platform. The example takes an RNA sequence and applies a quadratic model in pursuit of the optimal stem configuration.
> Jupyter Notebook
Scientific background: Fox DM, MacDermaid CM, Schreij AMA, Zwierzyna M, Walker RC. RNA folding using quantum computers,. PLoS Comput Biol. 2022 Apr 11;18(4):e1010032. doi: 10.1371/journal.pcbi.1010032. PMID: 35404931; PMCID: PMC9022793
Enzyme-Target Prediction on the Dynex Platform
The Dynex SDK based program predicts potential interactions between enzymes and target molecules and leverages the principles of quantum mechanics.

> Jupyter Notebook
Scientific background: Hoang M Ngo, My T Thai, Tamer Kahveci, QuTIE: Quantum optimization for Target Identification by Enzymes, Bioinformatics Advances, 2023;, vbad112
Efficient Exploration of Phenol Derivatives with Dynex
Molecule screening from a vast number of possible compounds is a challenging task. The emergence of quadratic unconstrained binary optimization (QUBO) solvers provides alternatives to address this issue. We propose a process for screening molecules by integrating QUBO solvers and density functional theory (DFT) calculations. As a proof-of-concept work, we map the problem of screening phenolic inhibitors onto the QUBO model. We approximate the bond dissociation energy (BDE) of the −OH bond, an indicator of good polymeric inhibitors, into the QUBO model by modifying the group contribution method (GCM) with the aid of DFT calculations. We demonstrate a strong correlation between this QUBO model and the data from DFT, with the correlation coefficient and Spearman’s coefficient of 0.82 and 0.86, respectively, when tested on the 85 given molecules. This mapping allows us to identify the candidates through the QUBO solver, whose BDEs are validated through DFT calculations, as well. Our work provides a promising direction for incorporating the GCM into QUBO solvers to tackle the molecule screening problems.

Scientific background: Efficient Exploration of Phenol Derivatives Using QUBO Solvers with Group Contribution-Based Approaches; Chien-Hung Cho, Jheng-Wei Su, Lien-Po Yu, Ching-Ray Chang, Pin-Hong Chen, Tzu-Wei Lin, Shin-Hong Liu, Tsung-Hui Li, and Ying-Yuan Lee; Industrial & Engineering Chemistry Research 2024 63 (10), 4248-4256; DOI: 10.1021/acs.iecr.3c03331
Breast Cancer Prediction
This examples shows using the Dynex SDK Scikit package which provides a scikit-learn transformer for feature selection using the Dynex Neuromorphic Computing Platform. The number of features have impact on neural network training and accuracy. It will be demonstrated how a significant reduction of features lead to similar (or even better) results.
> Jupyter Notebook
Scientific background: Bhatia, H.S., Phillipson, F. (2021). Performance Analysis of Support Vector Machine Implementations on the D-Wave Quantum Annealer. In: Paszynski, M., Kranzlmüller, D., Krzhizhanovskaya, V.V., Dongarra, J.J., Sloot, P.M.A. (eds) Computational Science – ICCS 2021. ICCS 2021. Lecture Notes in Computer Science(), vol 12747. Springer, Cham
> Automotive, Aerospace, Super-Sports and Space

Quantum computing is on the verge of transforming the pharmaceutical industry by vastly improving the speed and accuracy at which companies can discover and develop new drugs. This cutting-edge technology offers the ability to simulate and analyze complex molecular interactions at a quantum level, something that traditional computing methods struggle to accomplish due to the sheer complexity of biological systems. By enabling more precise modeling of molecular structures and their interactions with biological targets, quantum computing can significantly reduce the time and cost associated with the drug discovery process, allowing for the rapid identification of promising compounds.
Furthermore, quantum computing has the potential to revolutionize personalized medicine. It can process vast datasets from genetic information, enabling the design of treatments that are tailored to individual genetic profiles, thereby enhancing their effectiveness and reducing side effects. The ability to quickly analyze and interpret large volumes of genetic data also accelerates the understanding of diseases at a molecular level, leading to breakthroughs in the treatment of complex conditions such as cancer, Alzheimer's, and heart disease.
In addition to drug discovery and personalized medicine, quantum computing can optimize manufacturing processes and supply chain logistics in the pharmaceutical industry. By identifying the most efficient pathways for chemical synthesis, quantum computing can help in the production of drugs at lower costs and with reduced environmental impact. Similarly, sophisticated quantum algorithms can improve supply chain efficiency, ensuring that medications are produced and distributed in the most effective manner possible.
As quantum computing technology matures and becomes more accessible, its integration into the pharmaceutical industry is poised to usher in a new era of medical innovation. The promise of faster, more efficient drug discovery and development, coupled with the potential for personalized treatments, positions quantum computing as a key driver of future advancements in healthcare and medicine, marking a significant leap forward in our ability to combat disease and improve human health.
Drug Repurposing With 3D Molecular Quantum Methods
From concept to treating a patient, it can take 10 years for a single treatment. Drug repositioning, repurposing, re-tasking, re-profiling or drug rescue is the process by which approved drugs are employed to treat a disease they were not initially intended/designed for. Virtual screening has become essential at the early stages of drug discovery. However, the process still typically takes a long time to execute since it generally relies on measuring chemical similarities among molecules. Even for today’s processors, this exercise comprises a major challenge since it is computationally heavy and expensive. Accordingly, most of the well-known methods typically use 2D molecular fingerprints to include structural information that represents substructural characteristics of molecules as vectors. These methods do not take into consideration relevant aspects of molecular structures such as 3D folding, although they are efficient in terms of execution times. At the expense of higher computing times, considering 3D structural properties of molecules substantially increases the accuracy of results. The 3D molecular Quantum method is computed efficiently on the Dynex platform and provides a superior virtual screening method.

Scientific background: Drug repurposing based on a quantum-inspired method versus classical fingerprinting uncovers potential antivirals against SARS-CoV-2, Jimenez-Guardeño JM, Ortega-Prieto AM, Menendez Moreno B, Maguire TJA, Richardson A, Diaz-Hernandez JI, et al. (2022); PLoS Comput Biol 18(7): e1010330

RNA Folding
Finds the optimal stem configuration of the RNA sequence from the HIV virus and the Tobacco Mild Green Mosaic Virus using the Dynex platform. The example takes an RNA sequence and applies a quadratic model in pursuit of the optimal stem configuration.
> Jupyter Notebook
Scientific background: Fox DM, MacDermaid CM, Schreij AMA, Zwierzyna M, Walker RC. RNA folding using quantum computers,. PLoS Comput Biol. 2022 Apr 11;18(4):e1010032. doi: 10.1371/journal.pcbi.1010032. PMID: 35404931; PMCID: PMC9022793
Enzyme-Target Prediction on the Dynex Platform
The Dynex SDK based program predicts potential interactions between enzymes and target molecules and leverages the principles of quantum mechanics.

> Jupyter Notebook
Scientific background: Hoang M Ngo, My T Thai, Tamer Kahveci, QuTIE: Quantum optimization for Target Identification by Enzymes, Bioinformatics Advances, 2023;, vbad112
Efficient Exploration of Phenol Derivatives with Dynex
Molecule screening from a vast number of possible compounds is a challenging task. The emergence of quadratic unconstrained binary optimization (QUBO) solvers provides alternatives to address this issue. We propose a process for screening molecules by integrating QUBO solvers and density functional theory (DFT) calculations. As a proof-of-concept work, we map the problem of screening phenolic inhibitors onto the QUBO model. We approximate the bond dissociation energy (BDE) of the −OH bond, an indicator of good polymeric inhibitors, into the QUBO model by modifying the group contribution method (GCM) with the aid of DFT calculations. We demonstrate a strong correlation between this QUBO model and the data from DFT, with the correlation coefficient and Spearman’s coefficient of 0.82 and 0.86, respectively, when tested on the 85 given molecules. This mapping allows us to identify the candidates through the QUBO solver, whose BDEs are validated through DFT calculations, as well. Our work provides a promising direction for incorporating the GCM into QUBO solvers to tackle the molecule screening problems.

Scientific background: Efficient Exploration of Phenol Derivatives Using QUBO Solvers with Group Contribution-Based Approaches; Chien-Hung Cho, Jheng-Wei Su, Lien-Po Yu, Ching-Ray Chang, Pin-Hong Chen, Tzu-Wei Lin, Shin-Hong Liu, Tsung-Hui Li, and Ying-Yuan Lee; Industrial & Engineering Chemistry Research 2024 63 (10), 4248-4256; DOI: 10.1021/acs.iecr.3c03331
Breast Cancer Prediction
This examples shows using the Dynex SDK Scikit package which provides a scikit-learn transformer for feature selection using the Dynex Neuromorphic Computing Platform. The number of features have impact on neural network training and accuracy. It will be demonstrated how a significant reduction of features lead to similar (or even better) results.
> Jupyter Notebook
Scientific background: Bhatia, H.S., Phillipson, F. (2021). Performance Analysis of Support Vector Machine Implementations on the D-Wave Quantum Annealer. In: Paszynski, M., Kranzlmüller, D., Krzhizhanovskaya, V.V., Dongarra, J.J., Sloot, P.M.A. (eds) Computational Science – ICCS 2021. ICCS 2021. Lecture Notes in Computer Science(), vol 12747. Springer, Cham
> Automotive, Aerospace, Super-Sports and Space